Our research focuses primarily on two groups of flagellates: Euglenida and Preaxostyla. We are also mapping the diversity of protists in environments using metabarcoding.
We are particularly interested in the diversity of protists in environments and evolution of their semi-autonomous organelles, namely highly reduced or even completely lost mitochondrion-like organelle of anaerobic Preaxostyla and secondary plastid of photosynthetic euglenids. Both of these areas of research provide insight into organelle origin and evolution of their structure, molecular biology, transport, targeting, biogenesis, genome composition, molecular genetics mechanisms and biochemical pathways.
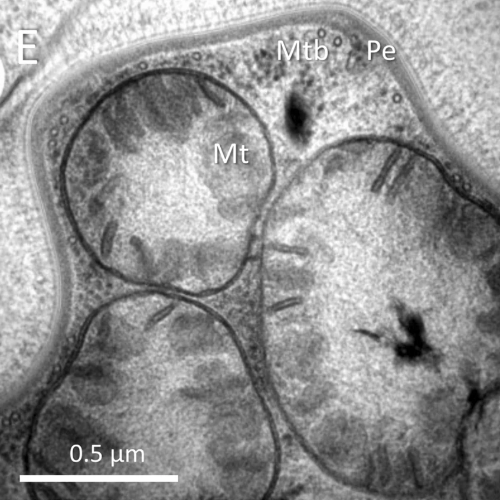

cells without mitochondria
Previously we demonstrated that the flagellate Monocercomonoides exilis (Oxymonada, Preaxostyla) does not have mitochondria. This is the fist such case among eukaryotes. We are interested in how such cells function, and in particular we are focusing on the process of synthesis of FeS centres, which is otherwise closely related to the mitochondria. We already know what genes are involved in this process, but we need to clarify many important details, e.g. where are the proteins localized in the cell, how they interact, what is the mechanism of FeS cluster assembly, what types of clusters are assembled, and finally, when the pathway evolved and who was the source of the genes.
We would also like to characterize the biochemistry of mitochondria in the cell of Paratrimastix pyriformis, a close relative of oxymonads, which offers a snapshot into the process of the loss of mitochondria in this group. To frame these events intophylogenetic context, we need to get a robust phylogenetic tree of the oxymonads, therefore we are hunting and sequencing oxymonads from the environments where they occur. Of interest are also metabolic interactions of some oxymonad species with their prokaryotic ecto- and endosymbionts.
Finally, we are using amitochondriate cells as models for "evolutionary" experiments. For example, we are interested in the evolution of import into mitochondria and we have proved that some proteins from amitochondriate cells are predisposed for import into mitochondria and mitochondrion-related organelles. We would like to understand which features are important in this phenomenon.
plastid of euglenids
It is clear that the green euglenid plastid was formed by secondary endosymbiosis with green algae. It is even known that this alga came from the evolutionary neighbourhood of the genus Pyramimonas. Since the first branches of the phylogenetic tree of green euglenids are composed of marine representatives, we assume that this event occurred in the sea. We do not know the age of this event, but it is clear that it is a younger endosymbiosis than most others.
In our research, we are interested in how the plastids of euglenids works. In particular, we are trying to find out what the origin of the plastid membranes is and how the proteins enter the organelle. These plastids have only three membranes compared to four as in the majority of secondary plastids, so which one has been lost? We have identified only a few proteins that might be involved in the protein import, but we have confirmation neither of their localization nor their function. Characterisation of RNAi knock-downs and CRISPRCas9 knock-outs may give us a hint.
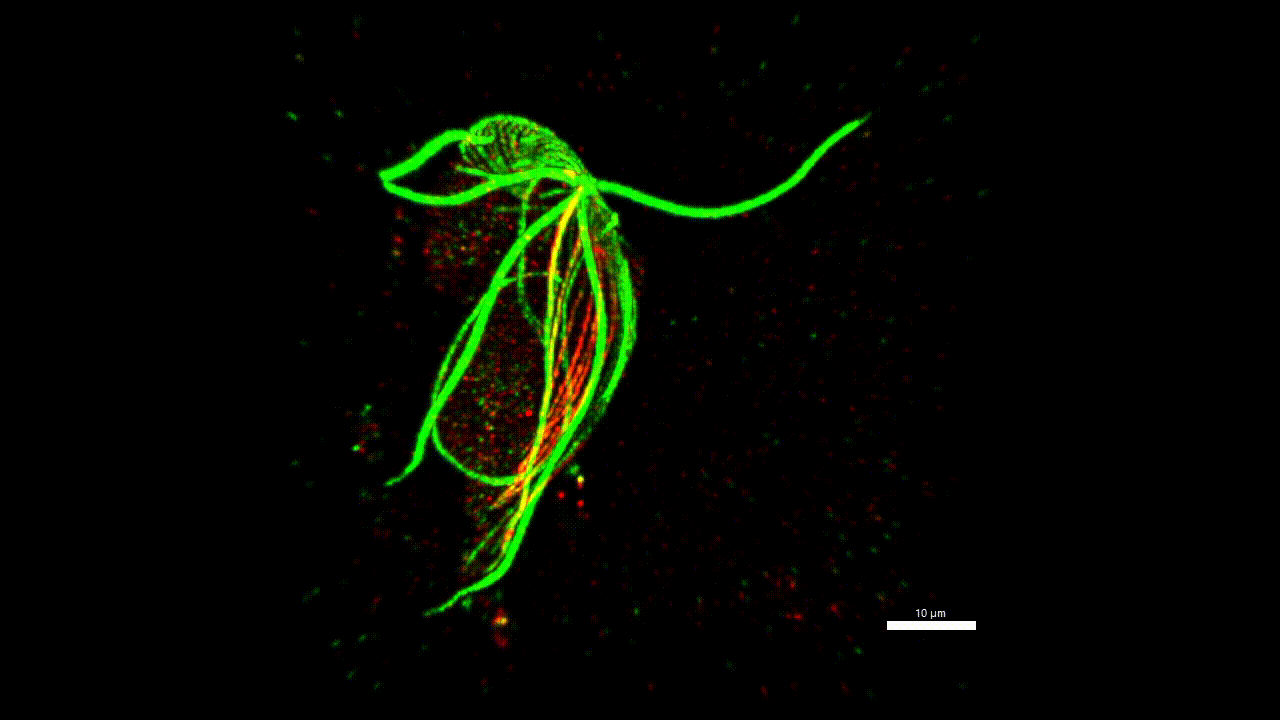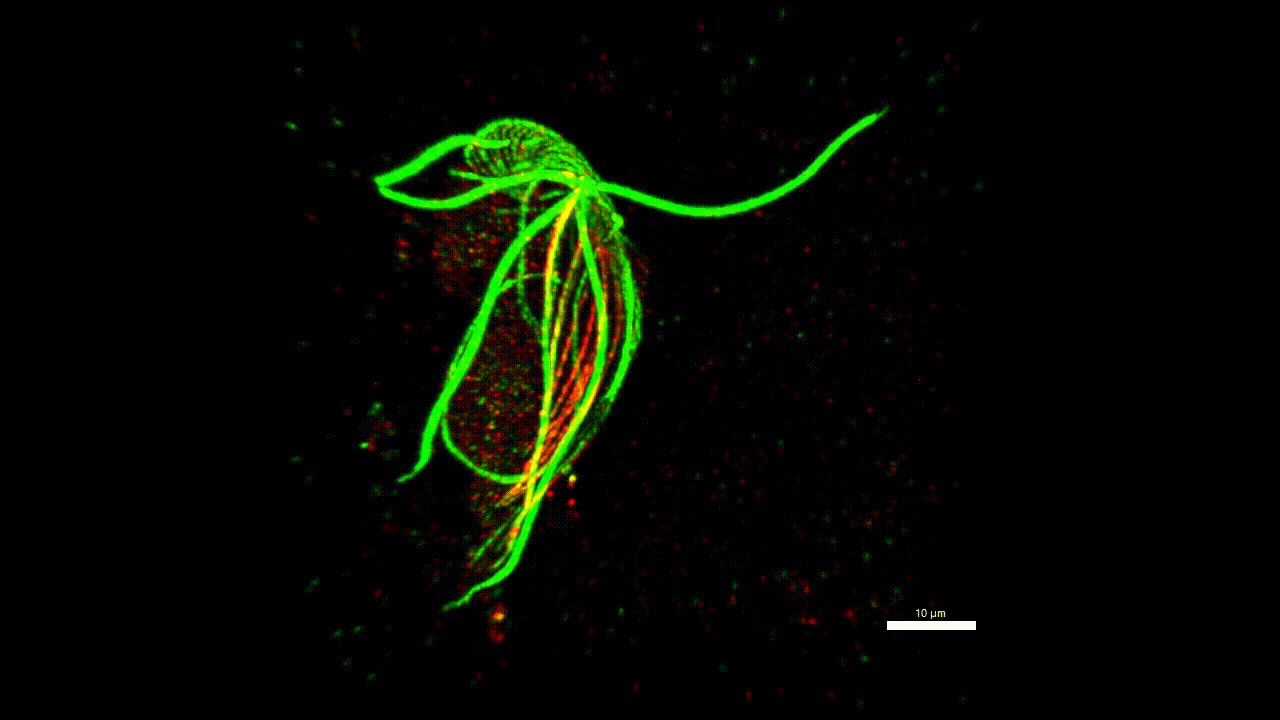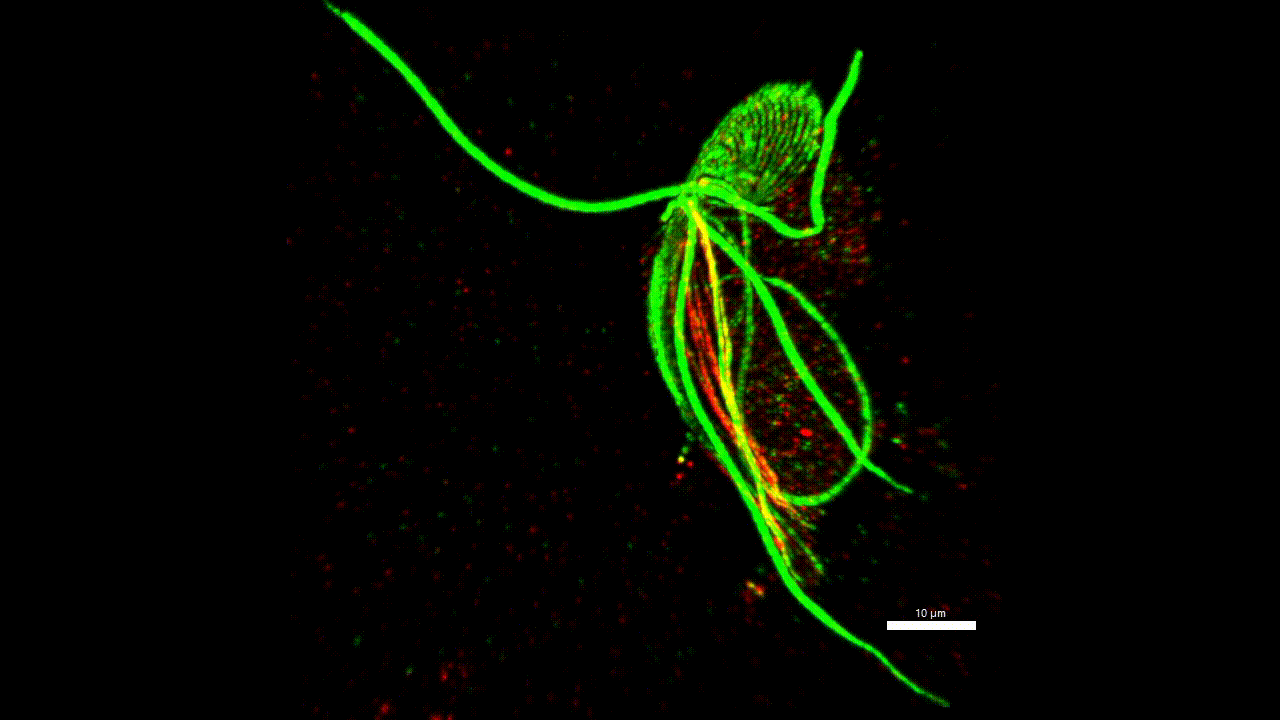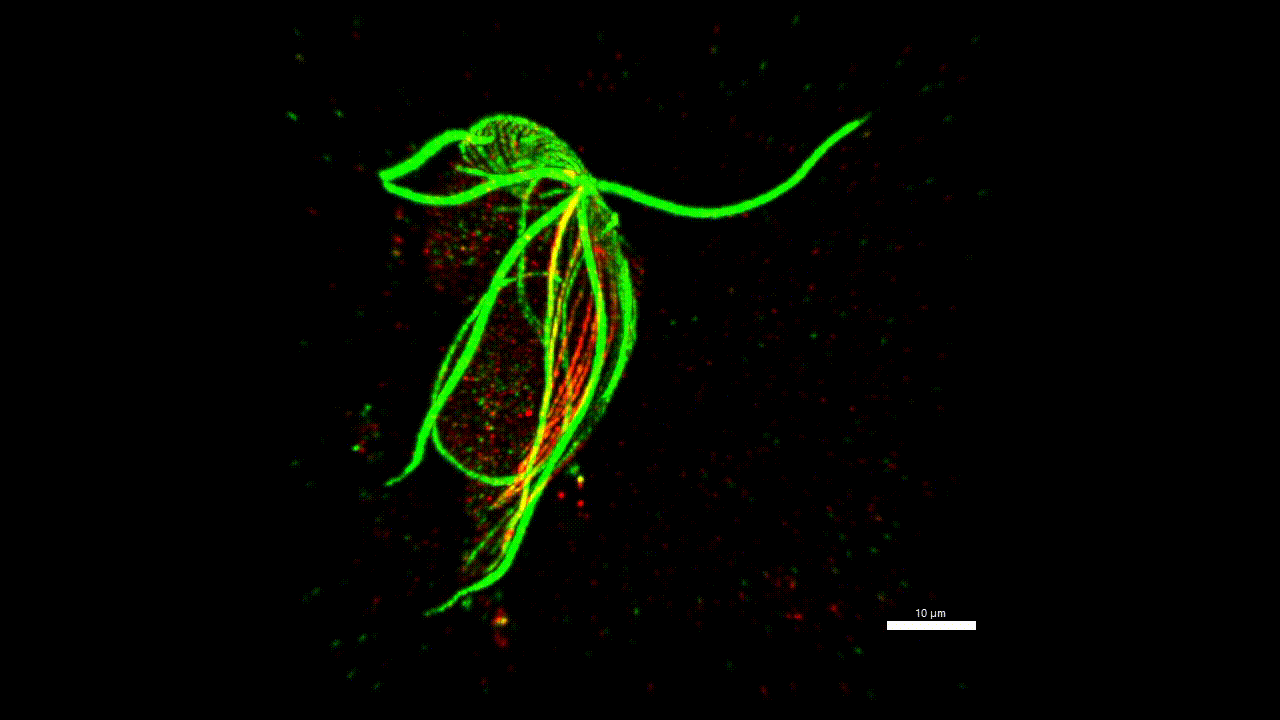
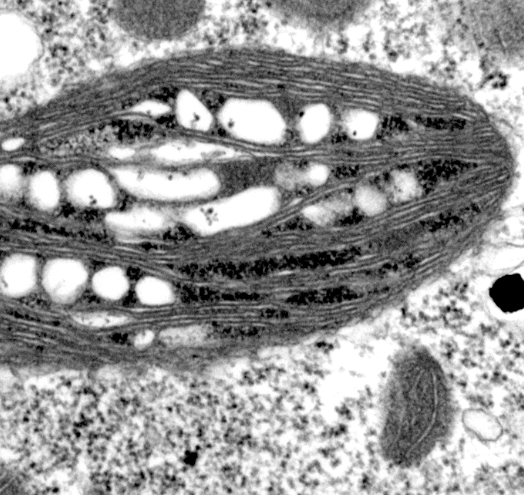
cytoskeleton of protist
Very little is known about the protein composition of the non-actin and non-tubulin cytoskeleton of protozoa, i.e. the different types of intermediate and annealed fibrils. Yet, this knowledge could allow us to homologize some morphological structures across distant groups and answer, for example, the question of what the ancestor of eukaryotes (LECA) looked like. We believe that the methodology has matured to a stage where it is possible to start asking such questions. Localisation of one new cytoskeletal protein (striated fiber assemblin) in Paratrimastix pyriformis suggests specific role in the formation of the excavate ventral groove (figure below). The elaborate structure beneath the surface of the euglenids (pellicles) gives the cells the ability for a special movement called "metaboly". Understanding the protein composition of this structure can help us understand the principle of this movement.